MEDtube is the largest social eLearning platform for 350,000+ professionals sharing 30,000+ videos, courses, images, documents and webinars
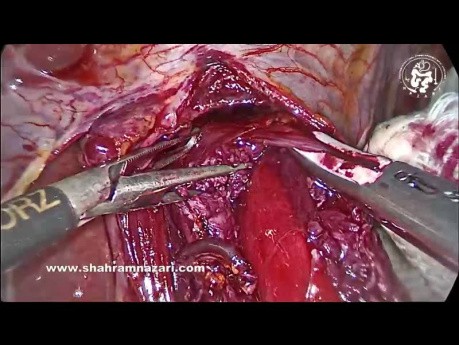
Register for freeTechnique of Heller Myotomy for Achalasia
Surgical video case: the easiest place to start the myotomy is on the distal esophagus 1-1.5 cm above GEJ. Grasp the 2 sides of the muscularis, folding the muscle into the jaws of the graspers. By streching...
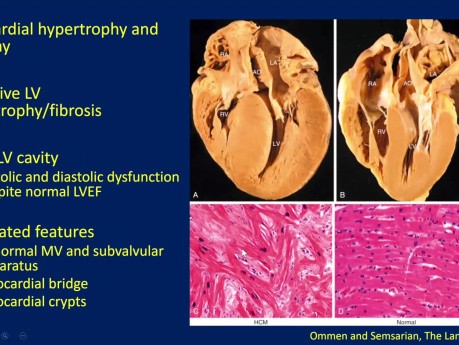
Echocardiographic Evaluation of Hypertrophic...
Echocardiographic evaluation of hypertrophic cardiomyopathy (HCM). Date: January 18, 2024 Speaker: Sean Cai MD FRCPC, Echo Fellow, St. Michael's Hospital. Objectives: 1. Understand basic pathophysiology...
Aortic Valve Disease Across the Ages
Aortic valve disease across the ages. Date: February 29, 2024. Speaker: Dr. Ra Han, Pediatric Cardiologist, St. Michael's Hospital Objectives: 1. Appreciate the impact of aortic valve stenosis on the...
Funky Motion: Basic and Advanced Wall Motion...
Funky motion: basic and advanced wall motion interpretation in echocardiography. Date: Thursday, 11 Jan 2024. Speaker: Howard Leong-Poi MD FRCPC Echocardiographer & Cardiologist, St. Michael's Hospital...
External Fixation
External Fixation (OrthoConcepts). Join the channel membership to unlock access to premium courses https://www. youtube.com/channel/UCpKGX6esbmV364XDaTQGckQ/join
Laparoscopic Common Bile Duct Exploration and...
Laparoscopic video case: this a cae of 52 year old female presented with abdominal pain and discomfort, laparoscopic cholecystectomy done 5 years back. Investigations revealed cbd calculi with concomitant...
Bronchial Wash
This video is presenting how to perform a bronchial wash with a single-use bronchoscope. The successful miniaturisation of Broncoflex has resulted in a large working channel in a medium-sized scope, supporting...
Donation After Cardiac Death
Professor Zeraatian's research on donation after cardiac death (DCD) illuminates a critical aspect of organ transplantation. His work delves into the ethical considerations, medical protocols, and logistical...
Chronic Rhinosinusitis with Polyps - Fullhouse...
Chronic Rhinosinusitis with Polyps - Fullhouse FESS. By Xavier Gonzalez-Compta MD, PhD at VII Hands on FESS course EENS Barcelona 2017.
Ectopic Pregnancy
Video case: 11 weeks pregnant lady completely asymptomatic came few days before her formal dating scan for just routine check up.
Biomechanics of Fracture Fixation
Biomechanics of fracture fixation. Join the channel membership to unlock access to premium courses https://www.youtube. com/channel/UCpKGX6esbmV364XDaTQGckQ/join
Featured channels
Mitral Valve Repair Center at The Mount...
As one of the world's most respected and experienced heart…
EUS - ENDO
EUS-ENDO will offer one day of live transmission from Paoli-Calmettes…
SAGES - Society of American Gastrointestinal...
The Society of American Gastrointestinal and Endoscopic Surgeons (SAGES) is…